It matters to know which PCV2 entered the herd

It looks like not all PCV2 viruses are created equal – science suggests that some appear to be more virulent than others. That is why it matters to understand what genotypes of the virus are around on a farm. Having exact insights allows a more targeted prevention campaign.
Porcine circovirus type 2 (PCV2) is a major swine pathogen, with wide distribution on most pig farms. A PCV2 infection is associated with various disease syndromes affecting pigs, especially in the grow-finish phase, denominated as Porcine Circovirus Diseases (PCVD). PCV2 has also been recognised as an important pathogen within the phenomenon called Porcine Respiratory Disease Complex.
Subdividing PCV2 into genotypes
Based on sequence identity in the capsid protein (the part of the virus that is exposed to the antibodies), PCV2 can be further subdivided into different genotypes. At the moment, scientists have described 9 of them. The PCV2 genotypes have been designated consecutively based on the time of first identification with lower case letters (’a’, ‘b’, ‘c’, until ‘i’). Different epidemiological and phylodynamic studies revealed the occurrence of different genotype shifts.
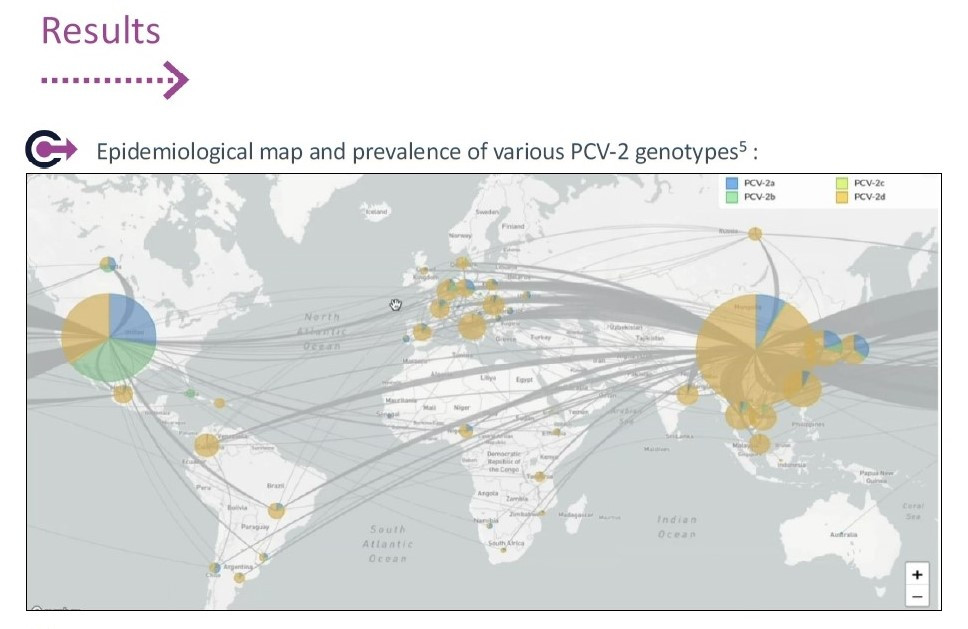
PCV2a was the most prevalent genotype in clinically affected pigs between 1996 and the 2000s, after which PCV2b predominated and was associated with the appearance of more severe clinical signs in disease outbreaks. Thereafter, a second shift from PCV2b to PCV2d occurred worldwide and was sometimes reported in cases of vaccination failure. PCV2d is nowadays most frequently or exclusively found, especially in clinically affected herds. Previously published data reported that 100% of isolates from diseased pigs were identified as PCV2d in Belgium, 100% in Spain and 60% in Austria. However, the detection of other PCV2 genotypes in vaccinated herds is still not unusual and, especially, when random sample selection is performed, PCV2a and PCV2b are still circulating in pig herds. A collection of results by Ceva in 2024 showed 100% of PCV2d in Spain, 87% in Poland, 80% in Denmark, 96% in Hungary, and 70% in the Benelux.
Those prevalence results are also reflected in public data, e.g., in the GenBank where laboratories from all around the world share genetic sequences; in particular, for complex clinical cases: since 2019, more than 88% of all reported PCV2 sequences are of genotype PCV2d, and this prevalence increases year after year.
Genotype and pathogenicity
There is some scientific controversy over the relationship between genotype membership and pathogenicity. Earlier studies indicated that PCV2b or PCV2d strains were more virulent and were responsible for potential vaccination failures. However, recent studies show that PCV2d virulence does not differ from PCV2a and PCV2b. Therefore, the experimental data obtained, so far, do not support a difference in virulence between the genotypes of PCV2. Nevertheless, differences in virulence were found in different mutants within the same genotype group. Moreover, a recent comparison of PCV2 genotypes a, b and d concluded that PCV2d was more virulent than PCV2a and PCV2b in pigs dually infected with PCV2 and M. hyo or with PCV2 and PRRS virus.
Risks of a co-infection
So far this article zoomed in on PCV2 alone. In case of a co-infection, like for instance with Mycoplasma hyopneumoniae, causative agent of enzootic pneumonia, pig health could be even more compromised. Yes, both pathogens do have different target organs and cells, yet the co-infection of PCV2 and M. hyo leads to more severe clinical respiratory disease, a lower weight gain, more extended lung lesions and an increased amount and prolonged presence of PCV2 antigens. It has also been demonstrated that M. hyo increased the severity of a subsequent PCV2 infection.
In addition, a double infection with both agents resulted in higher respiratory scores (especially for M. hyo and PCV2d) and in lower growth performance in pigs. M. hyo particularly increased the replication of PCV2d compared to other PCV2 genotypes. PCV2 and M. hyo infections also facilitate the expression of different secondary infections with the impact on overall morbidity and performance of pigs.
Prevention of PCVD and enzootic pneumonia is applied in most of swine farms worldwide by vaccination of piglets. The level of maternal immunity which might interfere with the post-vaccination response decreases usually for PCV2 and M. hyo rather rapidly. That allows vaccinating piglets against both agents at the same time, from three weeks of age, typically around weaning. Simultaneous vaccination against PCV2 and M. hyo is convenient, reduces the labour costs and is piglet-friendly. Single mono-valent, bi-valent ready-to-mix (RTM), or ready-to-use (RTU) vaccines are available in the market.
Combination vaccine with PCV2d antigen
In early 2025, Ceva Animal Health introduced the vaccine Cirbloc M. hyo, combining PCV2d and M. hyo antigens in a ready-to-use form. The M. hyo antigen is also being used in the Hyogen vaccine. The combined vaccine was successfully tested under both experimental and field conditions.
The relevance of a vaccine choice within the dominant PCV2d genotype can be assessed by a genotyping characterisation. For the Cirbloc M. hyo vaccine, 99% of all publicly available sequences of PCV2d since 2019 are very highly similar* to the vaccine capsid sequence.
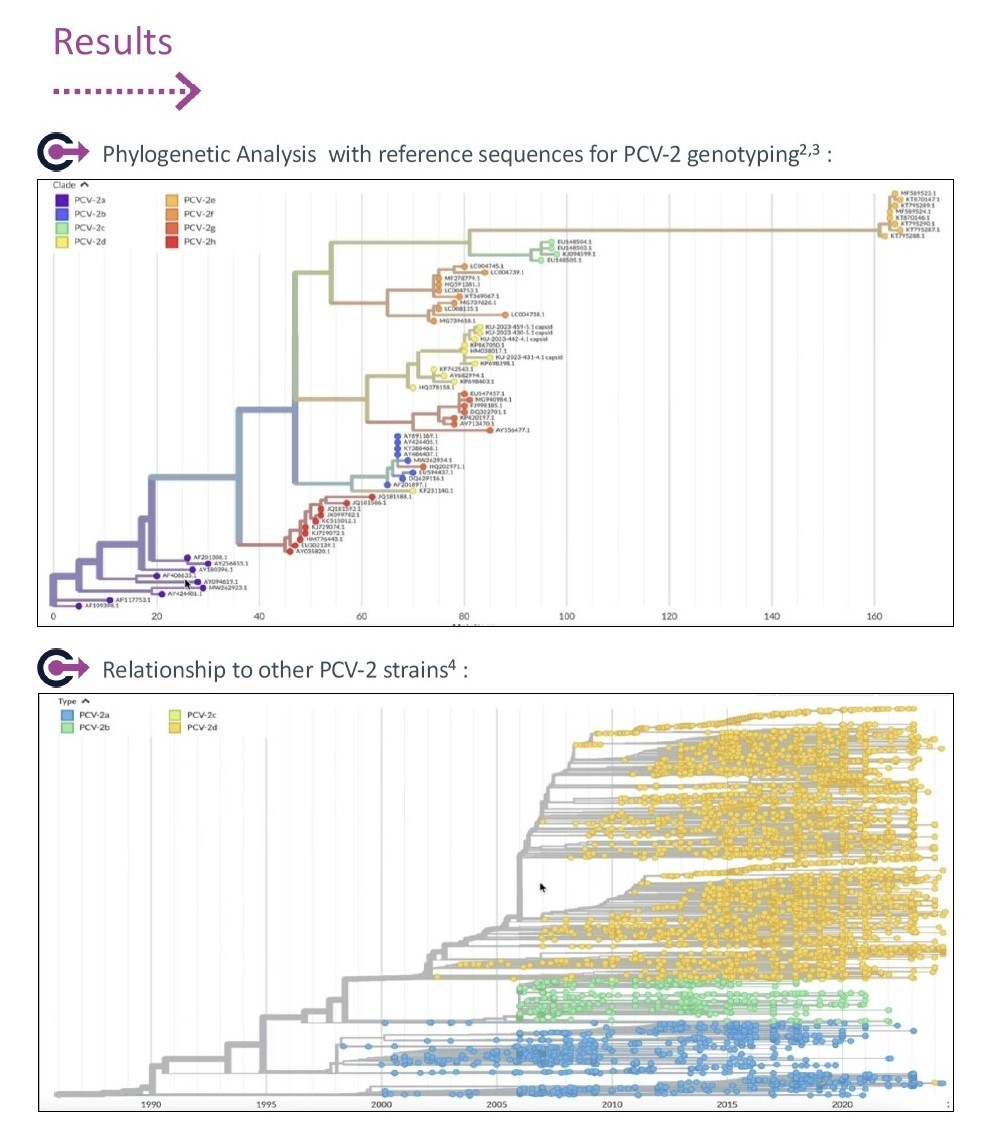
Genetic sequencing of PCV2 viruses
To better support concrete protection of the herds in the field, Ceva developed capacities to extract the genetic sequence of the virus through Next Generation Sequencing, and to compare it from a genomic and epidemiologic standpoint. To do so, the Nextstrain platform, originally developed to monitor human influenza and SARS-Cov-2, has been configured to received PCV2 sequences.
The genotyping of the virus from the capsid sequence has been derived from the reference for PCR tests using the Nextclade platform. Sequencing of the capsid protein of PCV2, thus, offers a more advanced view on virus properties and is nowadays more and more available as part of a diagnostic and monitoring approach. This is helping swine veterinarians and swine farmers making sure the vaccination programme in place on their farm is properly adapted to the epidemiology of the disease.
* Very highly similar is defined as being over 94% BLOSUM homology.